|
In
collaboration with
Prof.
Cees
Dekker at the
Kavli Institute of Nanoscience, Delft University of Technology*
Dr.
Keith Firman
at the University
of Portsmouth, and
Dr.
Mark Szczelkun
at the University of Bristol
(*This project started in the Molecular
Biophysics Group, Kavli Institute of Nanoscience, TU Delft)
References
L.K.
Stanley*, R. Seidel*,
C. van der Scheer, N.H. Dekker, M.D. Szczelkun & C. Dekker.
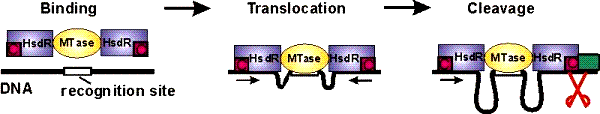
When a helicase is
not a helicase: dsDNA tracking by the motor protein EcoR124I
EMBO J. 25(10) 2230-2239 (2006) PDF Suppl. data The
EMBO Journal
*shared first authorship
R. Seidel,
J.G.P. Bloom,
J. van Noort, C.F. Dutta, N.H. Dekker, K.
Firman,
M. D. Szczelkun, and C. Dekker
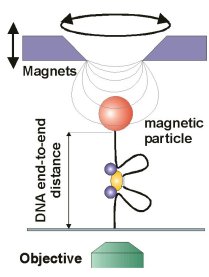
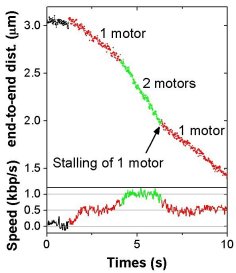
Dynamics
of initiation, termination and
reinitiation of DNA translocation by the motor protein EcoR124I
EMBO
J. 24(23),
4188-4197 (2005)
PDF
Suppl.
data The EMBO Journal
R.
Seidel, J. van
J. van
Noort, T.
van
der Heijden, C.
F. Dutta, K. Firman, and C. Dekker
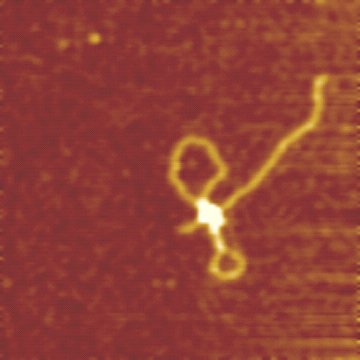
Initiation
of Translocation by Type I Restriction-Modification Enzymes
is Associated with a Short DNA Extrusion
Nucleic
Acids Res. 32(22),
6540-6547
(2004) PDF
Home
HT141230
Last
updated May 18, 2006
|